VarElect is a cutting edge Variant Election application for disease/phenotype-dependent gene variant prioritization. It is an effective and user friendly tool for analyzing genes with variants following Next-Generation Sequencing (“NGS”) experiments. VarElect can rapidly prioritize genes that have been found to have variants according to selected disease/phenotype - gene associations.
VarElect leverages the rich information within the LifeMap Knowledgebase of GeneCards®, the leading human gene database, MalaCards, the human disease database, LifeMap Discovery®, the regenerative medicine database, and PathCards, the unified human biological pathways database.
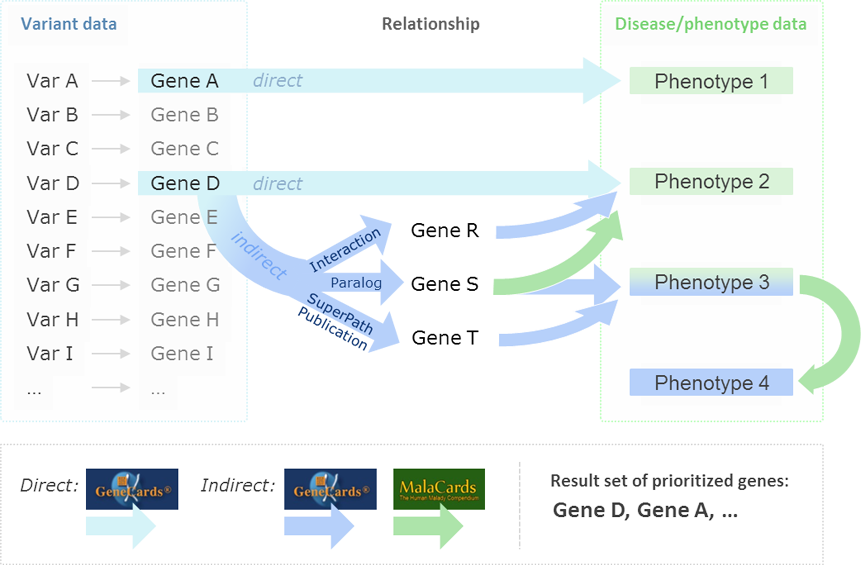
Using VarElect you can input a list of dozens, or hundreds of genes with variants, and narrow it down to the top 1-10 genes that are potentially associated with a particular disease, phenotype, or other biological function. VarElect acts jointly on the gene list and phenotype/disease keywords, and produces a list of prioritized, scored, and contextually annotated genes and direct links to supporting evidence and further information. VarElect utilizes the deep LifeMap Knowledgebase to infer direct, as well as indirect, links between genes and phenotypes. Indirect association between genes and disease are based on shared pathways, interaction networks, paralogy relations, domain-sharing, and mutual publications.
An example of an indirect GeneCards-based inference of GeneA to Phenotype PhenX is when PhenX is found to be associated with GeneB, which in turn shares a pathway with GeneA. Such gene-to-gene relationships are also formed (among others) by interaction networks, paralogy relations, domain-sharing, and mutual publications. MalaCards, in turn, allows one to produce a comprehensive phenotype search expression by using its data about diseases, underlying symptoms, and their relationships. The degree of mutual linking is quantified via endogenous search scores. VarElect provides a robust algorithm for ranking genes within a short list, and pointing out their likelihood to be related to a disease, enabling the researcher to perform the last decision step in deep sequencing runs in a fast and objective manner.
VarElect queries the GeneCards Suite Integrated Biomedical Knowledgebase (which includes GeneCards, MalaCards and LifeMap Discovery) for the specified phenotypes, and scores each hit according to its relevance.
Search query analyses - VarElect supports boolean expressions (using AND, OR, NOT and parenthesis, in addition to quotation marks for exact matches) and finds all genes meeting the criteria in the search query. Boolean expression examples –
Genes receive a relevance score depending (in part) on the weight of each query term that appears in relation to a given gene. The weight of a term is determined by the frequency it appears in association with a gene (term frequency) compared to all genes (inverse document frequency). If a term appears more often in the annotations associated with a given gene, and less often in all genes, the weight of that term for the given gene increases.
Boosting factors are applied to important fields (e.g disorders are boosted by 2). In addition, scores of all field hits are added together.
Finding relevant hits - VarElect first finds genes in the input gene symbols list that are directly related to the input phenotypes, and displays those “hits” in the “Directly Related” tab. Examples include genes where the phenotypes appear in one or more of the publications or disorders associated with the gene. Scores are calculated as described above.
The symbols of the genes that were not found to be directly related to the input phenotypes are queried to determine indirect connections to the phenotypes via intermediate genes. For example, a candidate gene, carrying a suspected variant as determined by Next Generation Sequencing, which is not directly connected to the phenotype of interest, may share a pathway with an implicating gene which IS directly connected to the phenotype; this establishes an indirect connection. Scores are given for implicating genes in relation to both an implicated gene (from the NGS experiment) and the phenotype. The geometric sum of the scores of the implicating genes of each implicated gene is calculated.
Minicards display the evidence used to connect a gene from an NGS experiment with the phenotypes directly or for the implicating genes which connect it indirectly.
Our search engine is based on Elastic Search. For more information about Elastic Search, please refer to elastic search documentation at (http://www.elasticsearch.org).
VarElect allows phenotype interpretation for non-coding variants sequenced by Whole Genome Sequencing (WGS). This functionality is implemented by leveraging GeneHancer - the regulatory elements (promoters and enhancers) database of the GeneCards Suite (more details in the GeneHancer paper and the GeneCards Guide). In addition to gene symbols, VarElect input can include variant-containing regulatory elements (called GeneHancers), relate them to target genes and include those genes in the VarElect analysis along with variant-containing genes.
After mapping variants of interest to GeneHancers, GeneHancer identifiers (GHids) can be specified in the Gene/GeneHancer input box. In the interpretation results page, genes inferred via GeneHancers are annotated with all genes, marked by the relevant GeneHancer identifier (symbol::GHid).
When analyzing GeneHancers, the phenotype score is calculated as follows. Each gene-GeneHancer association has a total score, calculated by multiplying the GeneHancer confidence score by the GeneHancer–Gene association score. Total scores are normalized to a range of 0.05-0.8, and each gene-phenotype score of the GeneHancer gene target is multiplied by the normalized GeneHancer total score.
Please contact us for a GeneHancer data dump.