Rapid prioritization of variant genes based on disease/phenotype of interest
SIGN UP FREEPowered by LifeMap's GeneCards Suite and its extensive phenotype - gene relationships annotations.
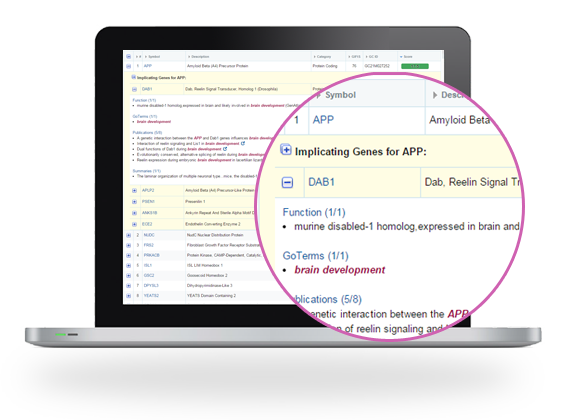
Unlike other platforms, leverages unique, extensive data from the LifeMap Integrated Knowledgebase to infer direct, as well as indirect links, between genes and diseases/phenotypes.
Provides valuable results without the need for complex bioinformatics expertise or tools.
Powered by LifeMap's integrated knowledgebase, which utilizes data from more than 100 select sources.
Provides annotated results with direct links to supporting evidence and further information available in the LifeMap Integrated Knowledgebase.
Our algorithms utilize associations between genes and diseases/phenotypes based on shared pathways, interaction networks, paralogy relations and mutual publications.
Stelzer G, Plaschkes I, Oz-Levi D, Alkelai A, Olender T, Zimmerman S, Twik M., Belinky F, Fishilevich S, Nudel R, Guan-Golan Y, Warshawsky D, Dahary D, Kohn A, Mazor Y, Kaplan S, Iny Stein T, Baris H, Rappaport N, Safran M, and Lancet D
VarElect: the phenotype-based variation prioritizer of the GeneCards suite,
BMC Genomics (2016), 17(Suppl 2):444; DOI: 10.1186/s12864-016-2722-2. [PDF]
Stelzer G, Rosen R, Plaschkes I, Zimmerman S, Twik M, Fishilevich S, Iny Stein T, Nudel R, Lieder I, Mazor Y, Kaplan S, Dahary, D, Warshawsky D, Guan-Golan Y, Kohn A, Rappaport N, Safran M, and Lancet D.
The GeneCards Suite: From Gene Data Mining to Disease Genome Sequence Analysis,
Current Protocols in Bioinformatics (2016), 54:1.30.1-1.30.33. doi: 10.1002/cpbi.5. [PDF]